Jonathan McCabe has always created fascinating examples of algorithmic art.
His paper Cyclic Symmetric Mutli-Scale Turing Patterns explains a process that creates very interesting looking images like the following.

With some help from a very patient Jonathan (many thanks Jonathan) he explained the process in more detail so I was able to implement the algorithms myself and experiment.
If you want to play with these patterns they are now included in Visions Of Chaos.
Explanation
Here is my attempted explanation of how the process works – aimed more at the programmer who would like to try implementing it themselves.
The process is based on a series of different turing scales. Each scale has values for activator radius, inhibitor radius, a small amount, weight and symmetry. These are the settings that control the results. For example

Create a main grid 2D array.
Create 3D arrays for activator, inhibitor and variations. In my case I used the format [x,y,scalenum] so each scale has its own dimension within the arrays.
Initialise the grid array to be the size of the image you wish to create and fill it with random floating point values between -1 and +1.
During each step of the simulation;
For each of the mutlitple turing scales;
1. Average each grid cell location over a circular radius specified by activator radius and store the result in the activator array. Multiply by the weight.
2. Average each grid cell location over a circular radius specified by inhibitor radius and store the result in the inhibitor array. Multiply by the weight.
3. Once the averages have been calculated, you need to work out the variation at each grid location. Within a specified variaition radius total up the variation using variation=variation+abs(activator[x,y]-inhibitor[x,y]). Store the variaition for that cell in the variation array. Remember each turing scale needs its own variation array. Smaller variation radii work the best. Checking only a single pixel for the variation formula gives the sharpest most detailed images.
Once you finally have all the activator, inhibitor and variation values for each scale calculated the grid cells can be updated. This is done by;
1. Find which of the scales has the smallest variation value. ie find which scale has the lowest variation[x,y,scalenum] value and call this bestvariation
2. Using the scale with the smallest variation, update the grid value
if activator[x,y,bestvariation]>inhibitor[x,y,bestvariation] then
grid[x,y]:=grid[x,y]+smallamounts[bestvariation]
else
grid[x,y]:=grid[x,y]-smallamounts[bestvariation]
The above step is the main gist of the algorithm. Update a single set of values based on multiple potential scales. That leads to the turing like patterns existing at the different scales within the single image.
Normalise the entire grid values to scale them back to between -1 and +1. ie find the maximum and minimum values across the grid and then update each cell
grid[x,y]:=(grid[x,y]-smallest)/range*2-1;
The grid values can now be mapped to a grayscale value (RGB intensities calculated by trunc((grid[x,y]+1)/2*255)). Alternatively you can map the value into a color palette.
Repeat the process as long as required.
Other Tips
To get images that look like Jonathan’s, you need to have a large difference of activator and inhibitor radius between the different scales. For example

The “Small amount” values really do need to be small. Especially when creating a movie. Otherwise the changes are too rapid and you lose that calm smooth slowly moving effect.
Speedups
After profiling the code, the slowest part of each step is calculating the averaged activator and inhibitor cell values. If using a circular radius then there is a sqrt and two sqr calls to see if the neighbour cell is within the specified radius. The sqrt can be skipped by comparing to the distance squared, but doing this for each scales cells really has an impact on perfromance. Be prepared to wait days if you are generating a movie at even a moderate size. Using the options above rendering a movie at 640×360 for YouTube is taking 27 minutes (!) per frame. All of this time (or 99.9% of it) is spent on the activator and inhibitor averaging, so I needed a much smarter way of doing this.
To help avoid all the distance calculations, you can create a list of neighbour cell offsets before going through each cell. That way when totalling each cells neighbours you use the currecnt cell position minus the saved offset coordinates. This helped speed up the code, but it was still too slow.
Next up was gaussian blur. Gaussian blurring can be simplified to do a pass over the horizontal lines and then the vertical lines of the arrays. This makes it much faster that the large circular averaging method and gave huge speedups (40 seconds per step compared to 27 minutes per step). The results are different to the circular neighbourhood method (as gaussian blurring uses a bell curve weighted kernel for the averaging) but the results are still acceptable.
Then after seeing this page with a fast 2 way rectangular blur I tried that. This gets the time down to 2.2 seconds per step. Using the rectangular neighbourhoods for blur can introduce small scale squared artifacts, but overall the results seem acceptable and the speed increase from 27 minutes per step down to 2.2 seconds per step is very welcome. You can apparently repeat the box blur 3 times to get results that approach a gaussian blur and it is still be faster than the usual gaussian blur. I tried this and it didn’t give the same as the gaussian, but it gets close and leads to more unique results.
Another speedup is related to the symmetries. When implementing the symmetry settings you need to average each of the activator and inhibitor values for each turing scale. This involves a lot of rotation calculations that need sin and cos calcs. It is much faster to precalculate each turing scales symmetry array locations once at the start and then use the precalced list during the main simulation steps. This got the time down to 780 milliseconds per step.
Since this blog post I also refactored the code to be multi-threaded where possible to take advantage of multi-core CPUs. This gives a big jump in performance on modern CPUs.
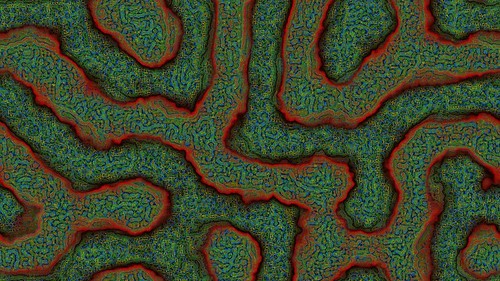
Result
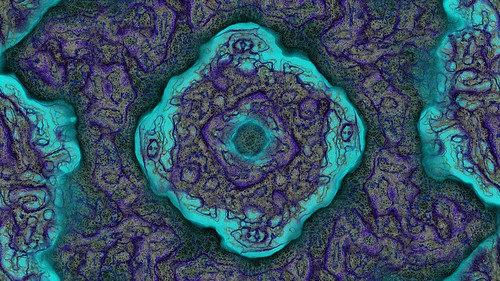
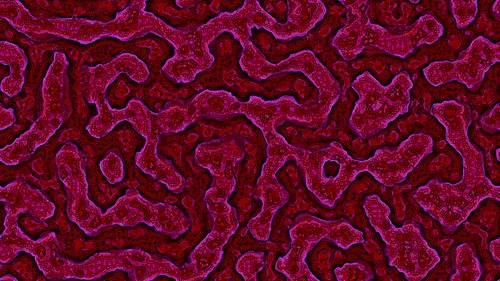
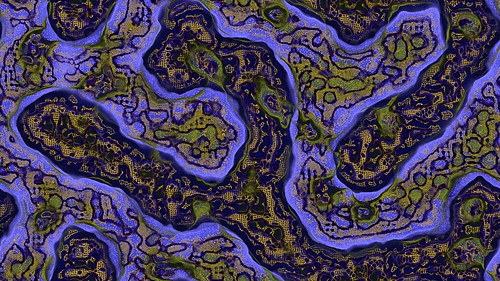
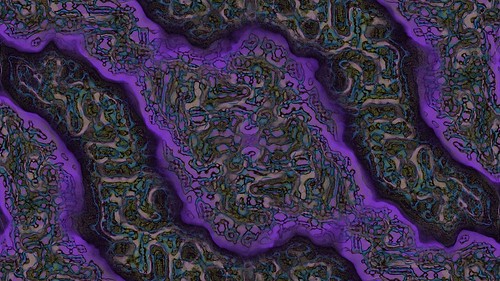
This is the result from using the settings shown above and the fast rectangular blur algorithm to average the activator and inhibitor neighbourhoods.
And here is another sample.
See here for more on multi-scale turing patterns.
Automatically searching for good looking results
Jonathan mentioned histograms in his paper and I have been trying to get my code “smart” enough to detect good or bad sets of random variables.
Each step of the simulation, map the grid cell values into a histogram. X axis is for cell values from -1 to +1. Y axis is the scaled percetage the number of times that cell value occurs.
“Boring” images will have a lot of low histogram counts. The following histogram was just prior to 99% of the values reaching the bottom and leading to a stabilised turing pattern.

The good looking images will have a wider variety of cell values even after many steps. These are the sort of histograms we are interested in detecting.

So once a new set of random variables is set, run the simulation for x number of steps. If most of the histogram values bottom out, then consider it bad and pick another set of variables and start again. If a specified number of steps is reached without a bad histogram, it is potentially good, so save the parameters with a thumbnail image and keep searching. This allows the computer to churn away overnight and look for good images hands free.
The trick is to tweak the bad histogram detection rates to not discard good results while still weeding out the bad results. At this stage I save both the bad and good results into separate folders to make sure it doesn’t discard any good images as bad.
Another method is mutation. Every 1 in 5 (or whatever ratio you like) load one of the good files and mutate the settings slightly. If the mutated image passes the histogram tests then save it with the other good finds.
After trying the histograms method for a while, it doesn’t really work. It will get rid of the most boring results, but between the detected good and bad rules there is no consistancy.
Another method I am trying is to detect how much the main grid array changes between steps. Set totalchange to 0 and then add each grid value totalchange:=totalchange+abs(grid[x,y]-oldgrid[x,y]). After all cells have been tallied you get a changefactor by totalchange/width*height. When auto-searching (after the first 25 steps for settle down) I consider the search bad if the changefactor is >0.01 or <0.006. This helps weed out a few more of the boring results, but there are still too many good results detected as bad and too many bad results detected as good.
So there needs to be smarter ways to try and find the best looking results without all the false-positives.
Adding Color
Make sure you click these images to see the full 4K resolution details.
I wanted to add color to the output images for a long time. Various methods I tried were not impressive (mapping the pixel brightness levels to a color palette or converting RGB to HSL all gave disappointing results).
Then I saw Ricky Reusser‘s twitter posts on how he had implemented and extended his Multi-scale Turing Pattern code to output color images. See his blog post here for more info.
To inject color into the Turing patterns the usual steps as described above are used except for a few small changes;
1. Before you start the steps, create a 2D array of colors that is the same size as the display image. Set all the values to the same starting color. I found black is a nice choice as the various colors fade in as the Turing steps run.
2. Set a different color for each of the scales. They can be any color, but brighter colors tend to work best.
3. At the point in the steps you find the bestvariation, you “bump” the current pixel color towards the scale color. For example if the best variation turns out to be the 2nd scale and that scale has a color of red RGB(255,0,0) you move the white color towards red. For the bump/move I used a simple linear interpolation (LERP).
target=trunc((current*(1-bumpamount))+(scalecolor*bumpamount))
This allows the colors to smoothly change as the steps of the simulation run.
4. To display the colors, you multiply the pixel color by the intensity of the grid value (grid[x,y]-1)/2.
That gives you the color shading. Some brightness and contrast tweaks can help bring out the colors a bit more.
This is also another example of the value of shared knowledge. Without this original blog post Ricky may never have seen these Turning patterns. Because of sharing the how to, he was able to come up with a new idea to extend the results further. I encourage everyone to share any knowledge you have to others.
Here is a sample movie of some color results.
More Color Ideas
Blake Jones has other coloring methods for turing patterns he describes here that I really need to experiment with when I get some time.
Jason.




Pingback: McCabeism: turning noise into a thing of beauty | W:Blut
Thank You for detailed explanations. This is very inspiring. I was wondered if it’s worth it to give it a try and implement in AS3, considering the heavy calculations, pixel manipulation and working with many massive stacks of arrays.
You’re welcome. Share the knowledge.
I have never used ActionScript myself, but give it a go. The basics of the algorithm are relatively simple and yet lead to the amazing cellular automata / reaction diffusion like results.
If you implement it, reply to this with a link.
Jason.
i had a crack at implementing this 2 weekends ago, good fun 🙂 basic animation: http://lyc.deviantart.com/art/Turing-patterns-245158514
i’ve yet to correctly implement the symmetry, and made a false start by trying to compute it in the final variance computation stage – presumably it’s done when computing the per-scale activation and inhibition buffers. i first converted the point in consideration to polar co-ordinates, and then averaged n samples (where n is the degree of symmetry) in a circle, with the 0th being the current point.
Yes, the symmetry is done (at least in my case which seems to work as expected) after you average the activator and inhibitor scales. So do the averaging, then do the symmetry on the averaged arrays, then use the symmetry averaged arrays to work out the variations.
If you pre-calculate and store the pixels for symmetry you can skip the calcs for polar conversion and rotation each step. For each activator and inhibitor radius, each pixel has a list of the other pixels it is symmeterised (sp?) with. Then when it comes time to do the symmetry you average each pixel with the other pixel coordinates in its list. If that makes any sense.
Jason.
Thanks for the clarification, I’ll report back once it’s working! Those images McCabe presented in his paper are really stunning, and I’d like to be able to get such results before improving the algorithm 😀
About the pre-computation, I can’t understand how it makes things faster. Modern computing is largely about reducing the impact of slow memory access – FLOPs are fast and memory accesses are extremely slow. (cf. http://www.akkadia.org/drepper/cpumemory.pdf, esp. page 16)
If I were going to make a lookup table to speed up the symmetry, it would just be a per-pixel polar co-ordinate conversion, to avoid having to do atan2 and sqrt. It may or may not be worth it, with memory accesses taking hundreds of cycles and sine/cosine being quite quick on modern hardware (even without SIMD optimisation). In fact, on a GPU I wouldn’t do any kind of precomputation at all, even going so far as to not make any buffers except the two strictly necessary for the current and next density grids, however there is local memory which can be exploited…
Can you please clarify the symmetry step ?
The way I understand it, is that (for example) for a symmetry of 4, we have 4 points on the circle around (0,0) with radius R, which are (R,0), (0,R), (-R,0), and (0,-R). So, the activator value in pixel (x,y) – let’s call it A(x,y), is replaced (at a given step) with:
A(x,y) = [A(x,y) + A(x+R,y) + A(x,y+R) + A(x-R,y) + A(x,y-R)] / 5
Is that correct?
Thanks.
Remember each pixel/point is rotated around the middle of the screen, not the origin.
Also for your example, you do not have 5 pixels, you take the current pixel and add the other 3 pixels at 90 degree increments. This gives you 4 in total to then average.
I used this code for doing the rotations
procedure Rotate2(RotAng: Double; x, y: Double; var Nx, Ny: Double);
var
SinVal: Double;
CosVal: Double;
begin
RotAng := RotAng * PI/180;
SinVal := Sin(RotAng);
CosVal := Cos(RotAng);
Nx := x * CosVal – y * SinVal;
Ny := y * CosVal + x * SinVal;
end;
procedure Rotate(RotAng: Double; x, y, ox, oy: Double; var Nx, Ny: Double);
begin
Rotate2(RotAng, x – ox, y – oy, Nx, Ny);
Nx := Nx + ox;
Ny := Ny + oy;
end;
So to find the other symmetry points for a pixel, use
rotate(360/n,x,y,xcenter,ycenter,xnew,ynew);
360/n is the degrees – n being the symmetry amount
x,y is the current pixel
xcenter and yceneter are the middle of the image
xnew,ynew are the returned new pixel locations
I also wrap-around the edges so if a retruned location is off the image it is wrapped around to the other side.
Hopefully that clears things up a bit?
Jason.
That’s clear, thanks! — I thought the rotation is around the pixel itself…
Hi Jason,
finally got some free time, and implemented in AS3. You can find it here:
http://esimov.zxq.net/lab/AS3/TuringPattern.html
Once again thanks for explanation.
Cheers, Endre
…and as I expected Actionscript is suffering at heavy calculation. With a smaller bitmap (much less pixels to compute) it’s working reasonably well, but scaling the dimension > 450×450 is struggling. It seems 200k pixels is almost the limit. Maybe some optimization would help a little bit.
Pingback: Organic Turing Pattern | hellopixel.net
I actually found out about this post from http://wonderfl.net/c/jmhz
An actionscript implementation and I added colour to it (not as sexy, http://wonderfl.net/c/sQHb)
I actually found this blog post via this site with an AS3 implementation which I then modified to give it colour.
http://wonderfl.net/c/jmhz – original
http://wonderfl.net/c/sQHb – modified
Pingback: Reaction-Diffusion Systems | Syntopia
basic implementation in WebGL shaders: check!
http://webglplayground.net/share/hrUMkUX5JR?fullscreen=0&width=1024&height=512&info=1&header=1
many regards!
I’ll add another framebuffer/texture loop for 4 more variations next.
(Gaussian blur radii are double per iteration)
Hi Jason,
Thanks for sharing your knowledge in such a clear way. I’ve implemented my own version with your directions and it’s doing some great things:
http://www.flickr.com/photos/vroegop/8051716789/ (4724 x 3272 at 12 seconds per frame :D)
http://youtu.be/P9IETXh64h4 (8 scales)
http://youtu.be/zkmM5J8NN6E (12 scales at full HD!)
I don’t know if I’ll be implementing the symmetry.
Many thanks!
JAV
Hi Jason,
Thanks so much for posting this. I read Jonathan’s paper and used this page as a guide to build a 3D version in Side Effects Software’s Houdini using only built-in nodes as part of a research project during my masters. I don’t currently have the files online, but you can check out an example in my demo reel (just before the minute mark):
where I use the resulting volume as a solid texture. It was a really fun project and I put a link to your blog in the credits list. thanks and keep up the great work!
kind regards,
alex
Hi Alex,
Very nice work with the 3D and texturing.
Regards,
Jason.
Very inspiring, Jason, thanks for that. I implemented a (surprisingly fast) Java applet with some coloring possibilities at http://maerschalks.byethost13.com/mathlab/jv_MSTP.html
(lots of examples and animations there)
Regards,
Piet
Hi Piet,
Nice job. It is fast for a Java applet.
Jason.
Hi, I am a student of computer science. I plan to implement the algorithm as a practice. There are a few points in the explanation part that I cannot understand.
Create 2D array for main grid is very intuitive.
I start to feel confused about “create 3D arrays for activator, inhibitor and variations”. Does that mean there are 3 different 3D arrays, for activator, inhibitor and variation respectively?
In the format [x,y,scalenum], does x,y means the coordinated of each grid cells? If this is a 3D array, does this means activator[x][y][scalenum], inhibitor[x][y][scalenum] and variation[x][y][scalenum]?
Hi Jacob,
Yes 3 different 3D arrays for activator, inhibitor and variation. Activator, inhibitor and inhibitor settings exist for a number of scales.
So, it is as you assumed. Activator[x,y,0] is the first activator scale XY coordinates. Inhibitor[x,y,1] is the second inhibitor scale array, etc.
Hope that clears it up.
Regards,
Jason.
Pingback: Multi-scale Turing Patterns – daily.hip
Pingback: Mesmerizing multi-scale Turing patterns in R with Rcpp | R-bloggers